Brain diseases, such as Alzheimer and Parkinson, severely affect the quality of life for a large number of people. The costs associated with medical and long-term care (in excess of 750 billion EUR in Europe in 2010) are rapidly rising, especially in relation to current/future trends of increased longevity and an aging population. Neuroimaging genetics is a young but quickly growing field, with enormous potential. The purpose of this VR project is to develop and define more reliable and powerful methods (statistics, image processing and machine learning) at the nexus of neuroimaging genetics, with the overall goal of exploring and explaining why and how specific brain diseases alter brain activity patterns, brain networks and behaviour.
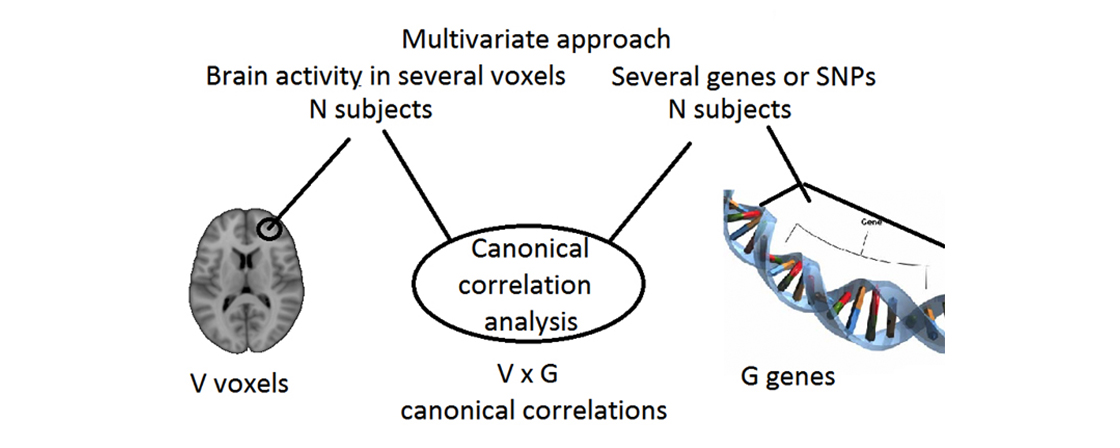
Functional magnetic resonance imaging (fMRI) can be used to study brain activity with high spatial resolution, and is based on the fact that the blood has different magnetic properties during rest and activity. fMRI can be divided into task fMRI (where the subject performs a specific task) and resting state fMRI (where the subject simply rests during the experiment, mainly used to study functional brain connectivity). The main application of diffusion magnetic resonance imaging (dMRI) is to study how nerve fibers connect different parts of the brain, and can thus be used to study structural brain connectivity. The method is based on measuring how easily water can travel in different directions. While fMRI and dMRI can be used to increase the knowledge regarding the human brain, and show functional or structural brain differences between healthy controls and subjects with a disease, the methods cannot be used to explain why these differences exist (or propose how to treat the diseases). It is therefore necessary to combine fMRI and dMRI with genetic data.
To reduce costs and save time, this project will not involve any data collection. We will instead use data from open repositories to develop and validate the new statistical methods. To collect fMRI and/or dMRI data from 100 subjects would cost 50,000 – 100,000 EUR and take 6 - 12 months. The Alzheimers disease neuroimaging initiative (ADNI) (http://adni.loni.usc.edu) shares neuroimaging (structural MRI, dMRI, resting state fMRI) and genetic data from 150 controls, 150 subjects with early mild cognitive impairment, 400 subjects with mild cognitive impairment, 150 subjects with late cognitive impairment and 200 subjects with Alzheimers (http://adni.loni.usc.edu/study-design/). The human connectome project (http://www.humanconnectome.org) shares neuroimaging and genetic data from 1,200 healthy adults. For each subject, resting state fMRI, task fMRI (for several tasks) and dMRI data are available. The Parkinson’s progression markers initiative (PPMI) (http://www.ppmi-info.org) shares neuroimaging (structural MRI, dMRI and resting state fMRI) and genetic data from 423 subjects with Parkinson and 196 healthy controls.
This project will focus on three aims, see Figure 1 for an overview.
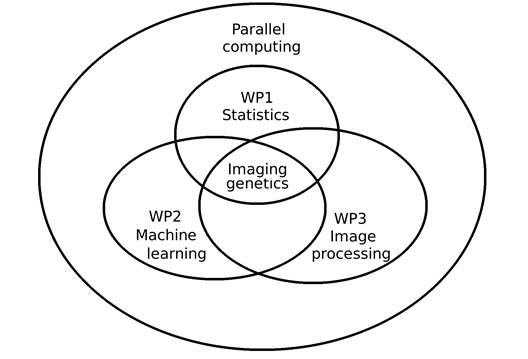
Figure 1. Overview of the work packages in the project. Parallel computing, e.g. using graphics cards, will be used in all work packages, due to the large datasets.
Aim 1 Increase the statistical power of neuroimaging genetics
To identify important risk factors for brain diseases, it is necessary to detect weak relationships between genes, brain function and brain structure. If a subject is healthy but has one or several risk factors, preventive measures can be applied to reduce the risk of developing a brain disease. A higher statistical power will be achieved using multivariate instead of univariate statistical algorithms (e.g. study relationships between several genes and several brain areas simultaneously), and by combining brain activity, brain connectivity, anatomical and genetic data (instead of only using two types of data).
Aim 2 Combine neuroimaging genetics with machine learning for early diagnosis of brain diseases
From a clinical perspective, it is more interesting to diagnose if a single subject has a brain disease or not, compared to doing large group studies. It is especially important to diagnose brain diseases early, to be able to apply different treatments, halt progression and thereby improve quality of life. Brain disease diagnosis will be achieved using machine learning techniques, which after training on a large number of patients and healthy controls can be used to classify whether a new patient has a brain disease or not. Inspired by aim 1, several types of data (representing brain activity, brain connectivity, brain structure, genetic information) will be combined to increase the sensitivity and the certainty of the diagnoses.
Aim 3 Improve the required preprocessing steps for neuroimaging genetics
Neuroimaging genetics studies are based on a large number of required preprocessing steps (mainly involving image processing), adding uncertainty to the final results. Erroneous relationships between genes and brain function may also be introduced. One of the required preprocessing steps is spatial normalization to a brain template, to be able to average brain activity or brain connectivity over subjects (brains). Other preprocessing steps include correcting for head motion and geometric distortions. These preprocessing steps will be improved using more advanced algorithms, to reduce the uncertainty.